Aquaculture Collaborative Research and Development Program (ACRDP) Fact Sheet
Issue 12 - May, 2012
Summary
Infectious Salmon Anemia virus (ISAv) poses a serious threat to fish farmers in Canada and globally. It can cause significant economic losses to Atlantic Salmon farming operations, and is a potential risk to wild Atlantic Salmon populations. Although improved farming and management practices have helped reduce the risk and impact of the disease, further outbreaks associated with new virulent strains remains a threat to the industry. A greater understanding of fish-virus interactions at the molecular level is of critical importance in the development of effective molecular tools and vaccines. A common approach for studying post-pathogen interactions at the molecular level is the use of functional genomic tools, such as DNA microarrays, which measure gene expression in specific tissues at specific times during the infection. With the use of this method, it has been possible to identify several genes up-regulated or down-regulated in salmon kidney tissue during the course of an ISAv infection and has provided a better understanding of how the fish fight the disease. Gene expression was measured at five different time points during an experimental infection: 6 hours, 24 h, 3 days, 7 d and 16 d after injection of the virus. Several genes involved in immune defense mechanisms were upregulated as the infection progressed. During the last stage of infection, several genes involved in oxygen transport were down-regulated. Such gene expression closely corresponds to anemia (low red blood cell count) observed in Atlantic Salmon infected by virulent strains of ISAv just prior to death. Such studies provide a deeper understanding of the defence mechanisms of fish against disease and provide insight into the approaches that could be taken to help the industry and prevent infections.
Introduction
Infectious Salmon Anemia virus (ISAv) poses a serious threat to stock health of the fish-farming industry. The virus can cause systemic disease that can lead to mass mortality in Atlantic Salmon (Salmo salar). Infected fish suffer from hemorrhaging, pale gills, congestion of internal organs and severe anemia, often leading to death. Other members of the salmonid family, including Brown Trout (Salmo trutta), Rainbow Trout (Oncorhynchus mykiss) and Coho Salmon (Oncorhynchus kisutch), can also be infected by ISAv. However, the majority of these infections are asymptomatic (a fish may carry the disease but experience no symptoms). Although ISAv has similarities with the influenza virus, it is not zoonotic (humans cannot be infected).
The disease was first reported in Norway in 1984 and has since been detected in Canada's East coast, the United States, Scotland, the Faroe Islands and Chile. The virus was identified in 1996 in New Brunswick, where it devastated the aquaculture industry. A number of biosecurity measures were introduced to limit the spread of the infection, such as aquaculture bay management units, but the risk of new outbreaks remains. Various strains of ISAv have been identified and characterized over the years and show a range of virulence from low to high, depending on the strain.
Although the evolution and pathogenicity of the various ISAv strains have been extensively documented, few studies have been conducted on the molecular interactions that take place between the virus and the host during an infection. When salmonids are infected with a virus, several defense mechanisms are activated, including an immune response and production of antibodies which will immunize the fish against subsequent viral attacks. However, with ISAv this response is often not sufficient to fight the virus, resulting in death of the fish. Understanding how salmon respond to the ISAv during the course of an infection is vital to the development of new antiviral treatments and new vaccines.
Given the major economic importance of salmon to the aquaculture industry and the potential disaster that could result from an ISAv infection outbreak, several teams of researchers are trying to develop a vaccine against the virus. When effective, vaccines stimulate the production of specific antibodies and protect the fish against future, related viral attacks. Effective vaccines are available for a number of fish diseases; Vibrio, furunculosis, etc., but to date, salmon have not responded well to the ISAv vaccines in development. The testing of vaccines is a long and costly process and therefore it would be extremely useful if a gene expression profile indicating the onset of the immune response in fish could be identified. This profile would be developed after vaccination and a short incubation period. It is very likely that effective vaccines would produce a genetic response in fish that is very similar to the response shown following an actual ISAv infection.
In recent years, a series of genome projects has led to the sequencing and construction of complementary deoxyribonucleic acid (cDNA) libraries for fish species, including Atlantic Salmon and Rainbow Trout. These cDNA libraries have made possible the development of DNA chips, which have been useful in many functional genomic studies. DNA chips consist of glass slides onto which DNA spots are printed. Each spot represents a unique genetic marker, usually a gene expressed in the host tissue.
The purpose of the present study is to identify genetic markers (a gene or DNA sequence of known location on a chromosome) indicative of an ISAv infection, as it progresses in time. It is assumed that many of these markers would also be present following vaccination with an effective vaccine formulation. An initial test of vaccine formulations could be performed to select the most promising for more detailed testing. The Consortium for Genomics Research on All Salmonids Project (cGrasp) developed a salmonid DNA microarray (or DNA chip) that holds 32,000 DNA spots. These DNA chips were usedto identify genes that were differentially expressed in the kidneys of Atlantic Salmon during the course of an infection with a virulent strain of ISAv.
Methods
Controlled exposure study
Prior to commencing a challenge, Atlantic Salmon smolts were housed for three weeks in sea water maintained at 13°C in the quarantine laboratory at the St. Andrews Biological Station, New Brunswick. The salmon were injected with a virulent isolate of the HPR4 strain of ISAv and held in duplicate tanks. The controls were injected with culture medium only. The fish were sampled at times 0 hours, 6 hours, 24 h, 3 days, 7 d and 16 d after injection and examined for signs of infectious anemia. Kidney sections were removed from each fish and preserved until analysis (Figure 1).


Figure 1
Sampling of Atlantic Salmon post injection with ISAv.Sample preparation
Total ribonucleic acid (RNA) was extracted from each kidney sample, purified, quantified and tested for quality control. Reference RNA was prepared from a separate group of healthy fish. The reference RNA represents the normal state of non-infected salmon. Each salmon sampled from the experimental tanks, injected with either ISAv or culture medium (control), was analyzed in comparison to the reference fish. For each sample of infected fish collected during the experiment, RNA was transcribed in complementary DNA (cDNA) and labeled (stained) in vitro with a fluorescent red dye. An RNA reference sample was similarly labeled, but with a green dye. Labeled cDNA from an experimental fish (control or infected) was mixed with cDNA from the reference fish. The mixture was then applied to the cDNA slide (microarray). Each of the 32,000 spots (or groups of identical DNA strands) on the slide represents a short gene sequence. When the slides are exposed to labeled cDNA from the infected and control fish, it hybridizes with the corresponding DNA spotted on the slides.
Scanning of DNA microarrays and image analysis
The DNA microarrays were laser-scanned and digitized for purposes of image analysis. The fluorescence intensity of each spot (gene) on the slide was measured. Fluorescence reflects the contribution of both fish (the cDNA of the reference fish, in green, and the cDNA of the experimental fish, in red). Since each spot on the slide represents a gene, the spots with a green intensity reflect a predominant contribution from the reference fish, or a reduced expression of that specific gene in the experimental fish. Spots with a red intensity reflect a marked predominance of cDNA from the experimental fish or an increased expression of that gene in the experimental fish. Fluorescence intensity indicates the amplitude of that differential expression. Genes that are not differentially expressed in the experimental fish will appear in yellow (Figure 2).
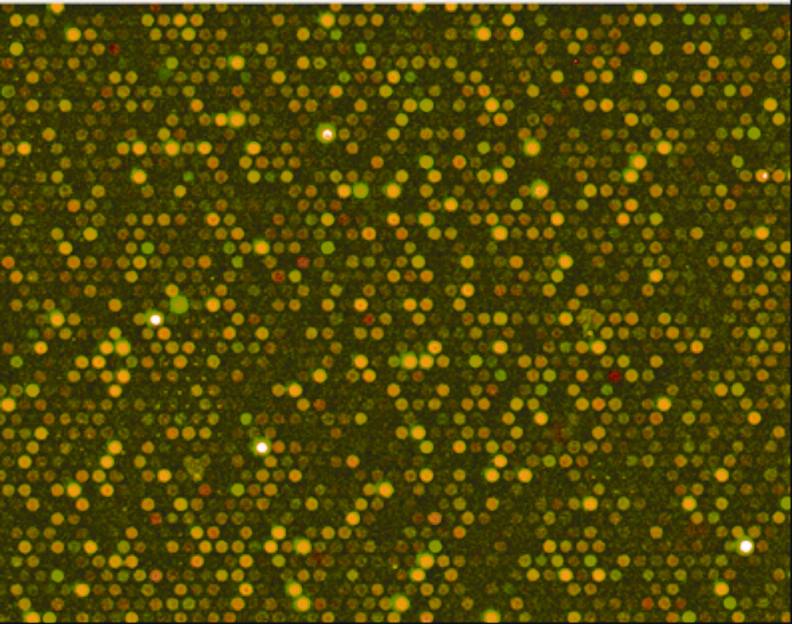
Figure 2
Gene mapping using fluorescence.Statistical analysis
Differentially expressed genes were identified for each sampling time point (6 hours, 24 h, 3 days, 7 d, 16 d) by comparing the ratios of ISAv-injected fish to fish injected with culture medium (controls). Significant genes identified for each sampling time point were then categorized by their function.
Validation by qRT-PCR
Quantitative Real-time reverse transcriptase polymerase chain reaction (qRT-PCR) was performed on each fish to verify the infectivity status. The same technique was used to confirm the differential expression of a selection of genes identified by the microarray analysis.
Results
The first mortalities of Atlantic Salmon infected by the ISA virus were observed on day 12 after the injection. On day 16, the end of the experiment, most of the remaining fish were close to death. The controls were healthy and tested negative for ISAv. The virus was detected in the kidney of ISAv-injected fish after 6 hours (3 out of 8 kidneys sampled). After 3 days, all injected fish tested positive and the viral load remained high for the rest of the experiment.
Several hundred genes were differentially expressed during the course of the infection, some observed after 6 hours and 24 h, but most observed at subsequent time points.
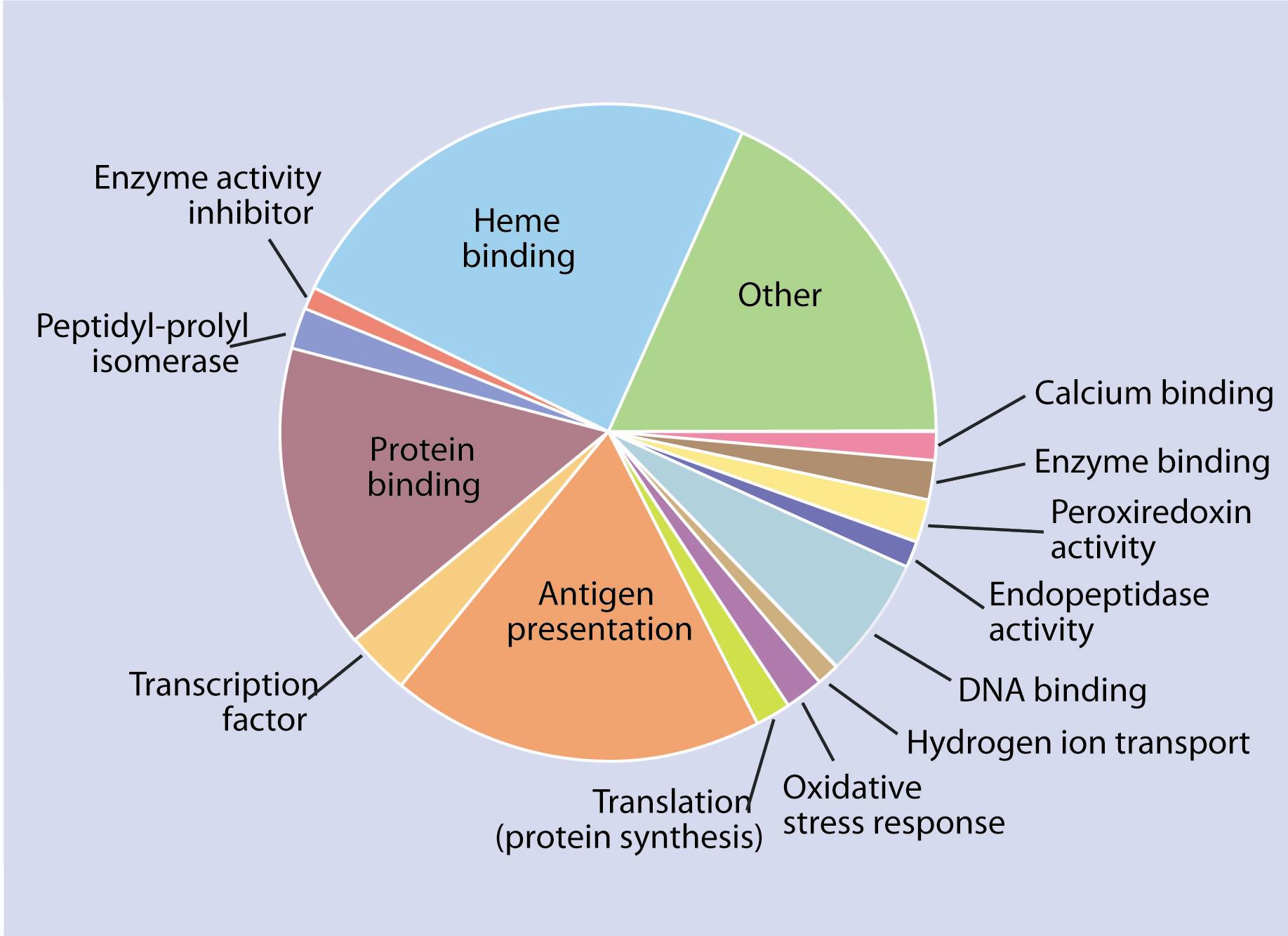
Figure 3
Proportion and function of genes expressed differentially after 16 days.Of those genes, a clear over expression of components associated with the development of the innate immune response and acquired immunity were observed. After 16 days, genes associated with an immune response (in this case antigen presentation to cytotoxic T lymphocytes - TCD8), were activated; several genes involved in oxygen transport were underexpressed (Figure 3).
As expected, overexpression of genes involved in the presentation of the virus to immune system cells was observed, but the activation of mechanisms leading to antibody production could not be detected. It is important to note that antibodies provide a long-term defence against subsequent infections. After 16 days, when most fish were dying, the viral infection had overcome the host's defence. Anemia (or blood deficiency) is a characteristic symptom of ISA infection and the ISA virus is known to adhere to the red blood cells of Atlantic Salmon. The gene response observed at that stage was closely correlated with the symptoms observed in the fish (i.e., underexpression of several elements associated with oxygen transport and red blood cell production).
From the list of genes identified as significantly expressed by microarray analysis, eight genes were selected and their expression confirmed using the qRT-PCR method (Figure 4).
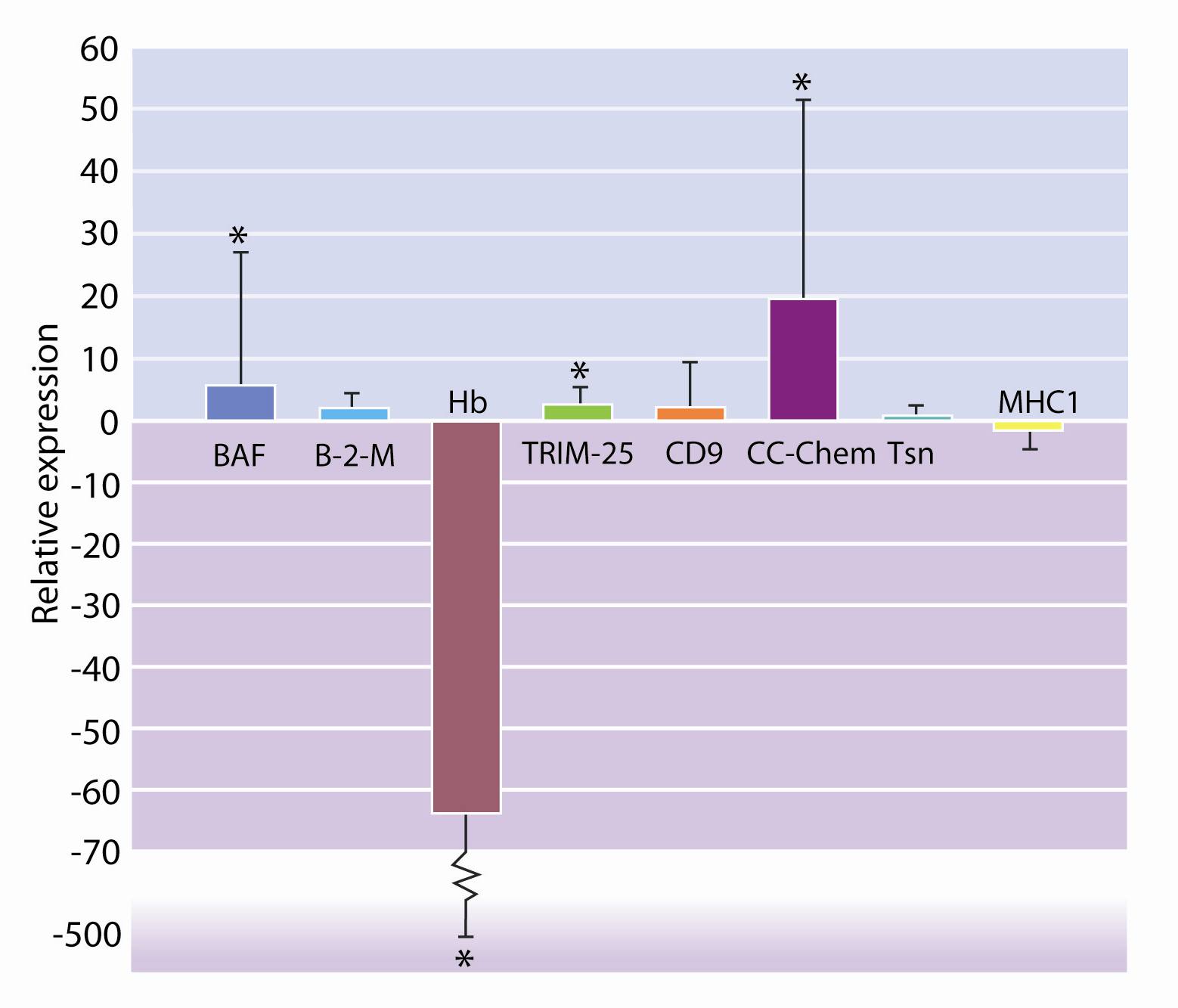
Figure 4
Relative expression of the genes selected.Conclusions
With the use of DNA microarrays, it was possible to detect the immune defence mechanisms of Atlantic Salmon to ISAv and to describe the sequence of physiological events leading to death. The genetic markers identified can be used to study new vaccines and to evaluate the effect of new isolates of ISAv. DNA microarrays are powerful tools for large-scale measurement of gene expression. The qRT-PCR method enables absolute quantification at a smaller scale and is a method of choice for validating the results obtained using microarrays. Once developed and validated using the qRT-PCR method, the results can be used in other similar experiments.
This experiment revealed that naïve salmon (salmon that have not been used in previous experiments) responded strongly to ISAv infection. It would be interesting to expand the research to include how other variables would be likely to affect the immune response in salmon (e.g., resistance of certain families of fish, the level of virulence of the isolate injected, environmental conditions that affect the disease such as hypoxia and temperature effects). Such studies provide a deeper understanding of the defence mechanisms of fish against disease and provide insights into the approaches that could be taken to help the industry and prevent infections.
This ACRDP project (MG-09-01-002) was a collaborative effort among the Fisheries and Oceans Canada (DFO), and the Atlantic Canada Fish Farmers Association. One of the scientists on this project, Nellie Gagné, can be contacted at Nellie.Gagne@dfo-mpo.gc.ca.
For further information on this and other ACRDP projects, visit: /aquaculture/acrdp-pcrda/index-eng.htm.
Published by:
Aquaculture Science Branch
Fisheries and Oceans Canada
Ottawa, Ontario K1A 0E6
©Her Majesty the Queen in Right of Canada 2012
ISSN 1919-6806 (Print)
ISSN 1919-6814 (Online)
ISBN: 978-1-100-20661-5
DFO/2012-1825
Cat No.: Fs23-580/3-2012E-PDF
French version and alternative formats available through: /aquaculture/acrdp-pcrda/index-fra.htm.
- Date modified: