Bonamia exitiosa (Bonamiasis of Various Oysters)
On this page
Category
Category 3 (Host Not in Canada)
Common, generally accepted names of the organism or disease agent
Microcell disease, Bonamiasis, Bonamiosis, Haemocyte disease of oysters.
Scientific name or taxonomic affiliation
Bonamia exitiosa (=exitiosus) (Hine et al. 2001, Berthe and Hine 2003) with close resemblance to Bonamia ostreae a pathogen of European flat oysters Ostrea edulis. Since the original description of this parasite, B. exitiosa and B. exitiosa-like microcells have been reported from various oysters in various locations around the world based on genetic similarities in the sequence of one gene, specifically, segments of the small subunit (SSU or 18S) and internal transcribed spacer regions (ITS1-5.8S-ITS2 which make up the ITS rDNA locus) of the ribosomal DNA gene complex (Hill et al. 2010b, Hill et al. 2014). Hill et al. (2014) recommended that the ITS rDNA locus should be used for determination of the species boundaries of B. exitiosa as well as other Bonamia species. Prior to 2010, it was not known whether the Bonamia exitiosa-like microcells consists of different strains of one species or the group consists of multiple species (Lormann et al. 2009, Hill et al. 2010b). For most of the microcells in this B. exitiosa-like group, relatively few data on histology, ultrastructure and molecular sequences was available and not sufficient to unequivocally discriminate between species (Hill et al. 2010b). Like other species of Bonamia, B. exitiosa was affiliated with the Haplosporidia based on DNA analysis (Hill et al. 2010b, Hartikainen et al. 2014). Engelsma et al. (2014) indicated that the genus Bonamia represents a derived clade within the phylum Haplosporidia whose members have generally adopted (1) life cycles based on direct oyster to oyster transmission of uninucleate amoeboid cell forms, and (2) intracellular infection of oyster haemocytes by these cell forms. Molecular analysis was used to distinguish two separate clusters within the genus Bonamia: a cluster with Bonamia perspora and Bonamia ostreae and a cluster of Bonamia exitiosa (-like) microcells (Hill et al. 2010b). Apparently, B. exitiosa and Bonamia roughleyi are genetically very similar (Abollo et al. 2008, Hill et al. 2010a). Hill et al. (2010a) grouped these two named species as the B. exitiosa/B. roughleyi clade and indicated that justification for drawing species boundaries among the primarily austral microcells with affinities to B. exitiosa and B. roughleyi remains elusive as confirmed by Carnegie et al. (2014).
Geographic distribution
Bonamia exitiosa was originally described in Ostrea chilensis from Foveaux Strait and other locations around South Island, New Zealand including Golden Bay, Tasman Bay, the Marlborough Sounds and Wellington Harbour (Dinamani et al. 1987, Hine 1997, Hine et al. 2001). More recently, B. exitiosa was possibly detected in Ostrea stentina (=Ostrea aupouria) from North Island, New Zealand (Hill et al. 2014). Since the late 1980s, unidentified Bonamia were reported from various locations around the world. Recent genetic analysis of one gene (specifically, segments of the small subunit (SSU or 18S) and internal transcribed spacer regions (ITS1-5.8S-ITS2) of the ribosomal DNA gene complex) was used to identify some of these organisms as B. exitiosa (White et al. 2008, Hill et al. 2014, Engelsma et al. 2014). The reported locations and corresponding hosts are as follows:
Australia: from New South Wales (various locations) in Ostrea angasi and possibly Saccostrea glomerata, (Corbeil et al. 2006b, Carnegie et al. 2014); and probably from Port Phillip Bay, Victoria, Georges Bay, Tasmania and Albany, Western Australia in O. angasi (Hine and Jones 1994, Engelsma and Hine 2009a, Hill et al. 2010b). While B. exitiosa was unambiguously detected using molecular methods in S. glomerata from southeastern Australia, visual evidence of infection remains elusive (Engelsma et al. 2014).
Europe: from the Galician coast (NW Spain), the Ebro Delta on the Mediterranean coast of Spain, the Manfredonia Gulf, Italy (Adriatic Sea) and southwest coast of England in Ostrea edulis including concurrent infections with Bonamia ostreae (Abollo et al. 2008, Narcisi et al. 2010, Carrasco et al. 2012, Ramilo et al. 2014b, Longshaw et al. 2013, Laing et al. 2014, Engelsma et al. 2014). Also, B. exitiosa was detected by molecular analysis in one larval sample from the pallial cavity of O. edulis collected in 2008 from the Bay of Quiberon, South Brittany, France (Arzul et al. 2011). Insinuations that B. exitiosa has been detected in O. edulis on the Mediterranean coast of France occur in Arzul et al. (2011) and in the OIE Manual of Diagnostic Tests for Infection with Bonamia exitiosa (2012).
Tunisia: from Hammamet in Ostrea stentina (Hill et al. 2010a, 2014).
USA: from North Carolina and Atlantic waters of southern Florida (Indian River Lagoon) in experimentally introduced Crassostrea ariakensis (Carnegie et al. 2005, 2006; Burreson et al. 2004; Bishop et al. 2006; Audemard et al. 2008a,b; Carnegie et al. 2008; Dungan et al. 2012); from North Carolina and South Carolina in the noncommercial oyster Ostrea stentina (=Ostrea equestris) which is also known to be infected by Bonamia perspora (Dungan et al. 2012, Audemard et al. 2014, Hill et al. 2014); and from California (Elkhorn Slough) in Ostrea lurida (Hill et al. 2014). Bonamia exitiosa may also occur in Crassostrea virginica in the eastern USA (Engelsma et al. 2014, Hill et al. 2014 and see OIE Reports 2012 & 2013 in References below). In situ hybridisation (ISH) was used to detect a parasite closely related to B. exitiosa in archived histological materials of O. edulis from Chincoteague Bay, Virginia (Hill et al. 2014).
Argentina: from San Antonio Bay, San Matías Gulf, Pategonia in Ostrea puelchana and Ostrea stentina (Kroeck and Montes 2005, Oehrens Kissner et al. 2014, Hill et al. 2014).
Genetically similar but unnamed parasites were reported from Ostrea chilensis in the Chiloe and Quihua Islands, Chile (Kern 1993, Campalans et al. 2000, Lohrmann et al. 2009). Further analysis by Hill et al. (2014) indicated that the Bonamia sp. in O. chilensis from Chile appeared in the B. exitiosa clade in SSU rDNA phylogenetic analyses, but diverged from this clade in the ITS rDNA analyses. This was also true for a closely related Bonamia sp. in Ostrea edulis from Tomales Bay, California, USA (Hill et al. 2014). Thus, Hill et al. (2014) and Engelsma et al. (2014) concluded that inclusion within B. exitiosa of the parasites from O. chilensis in Chile and O. edulis in California may not be justified, and these lineages may represent novel species.
Host species
Ostrea chilensis (=Tiostrea chilensis, =Tiostrea lutaria), Ostrea edulis, Ostrea angasi, Ostrea stentina (Ostrea (=Ostreola) equestris, =Ostrea aupouria), Ostrea lurida, Ostrea puelchana and Crassostrea ariakensis are hosts of B. exitiosa based on visualisation of the parasite in the oyster tissues and the similarity of the molecular sequence of regions of the ribosomal DNA gene complex (SSU rRNA operon and the ITS1 region) of Bonamia obtained from these oysters (Corbeil et al. 2006b, Abollo et al. 2008, Narcisi et al. 2010, Carrasco et al. 2012, Dungan et al. 2012, Longshaw et al. 2013, Ramilo et al. 2014b, Carnegie et al. 2014, Audemard et al. 2014, Hill et al. 2014). Crassostrea virginica may also be a suitable host (Engelsma et al. 2014 and see OIE Reports 2012 & 2013 in References below). However, the pathology associated with infection in O. chilensis and O. angasi is very different (Hine 1996b, Engelsma and Hine 2009a). In southeastern Australia, B. exitiosa was unambiguously detected using molecular methods in Saccostrea glomerata, however, visual evidence of infection remains elusive (Engelsma et al. 2014). Also, in Spain, B. exitiosa DNA (without visualizing the microcell) was detected in tissues of Crassostrea gigas highlighting the possibility of this parasite surviving extracellularly and in other non-typical hosts (Lynch et al. 2010). According to Hill et al. (2010b), examination of O. stentina (=O. aupouria) in New Zealand has produced a PCR product resembling B. exitiosa/B. roughleyi. Note that the oyster O. stentina (=O. aupouria, =O. equestris) is associated with B. exitiosa in New Zealand, Argentina, eastern USA, and the Mediterranean.
Impact on the host
Like Bonamia ostreae, B. exitiosa is an intrahaemocytic protozoan that quickly becomes systemic with overwhelming numbers of parasites coinciding with the death of the oyster. In New Zealand, large-scale mortalities of native dredge oyster (91% between 1975 and 1992 in Foveaux Strait (Doonan et al. 1994, Cranfield et al. 2005)) have been attributed to this parasite. The fishery was closed in 1993 with severe economic impact on local communities. Investigations have shown that B. exitiosa was present in Foveaux Strait oysters in 1964, and a long standing association between host and parasite is suggested by a consistent seasonal variation in infection (Hine 1991a,b). The well-defined annual pattern of infection consists of: very low levels of infection occuring in the austral winter, numerous small uni-nucleate stages being present in spring to early summer, larger amoeboid uni-nucleate stages occuring in late summer and autumn corresponding with the highest prevalence of infection, and the parasite population crashing in early winter (Hine 1991a,b). Analysis of the epizootic event from 1986-1992 lead Cranfield et al. (2005) to propose that stressors increased the susceptibility of the oysters to an enzootic parasite. In addition, epizootic mortalities have occurred among O. angasi in Tasmania, Australia, and in both New Zealand and Australia, attempts at farming oysters in infected areas have resulted in 100% mortalities (Engelsma and Hine 2009b). Bonamia exitiosa has emerged as the most serious disease threat for the introduced C. ariakensis on the east coast of the USA, especially under warm euhaline conditions and for oysters less than 50 mm in shell length. This disease threat may thwart the attempt to use C. ariakensis in efforts to build an aquaculture industry based on this oyster in the eastern USA (Audemard et al. 2014). In this area, the natural transmission processes of B. exitiosa displayed a stronger seasonal cycle with mortality peaks in the summer months (Carnegie et al. 2008) and the parasite being much less prevalent in winter (Engelsma et al. 2014).
On entry into the host, probably via the gut, B. exitiosa are phagocytosed by haemocytes but are not killed by these cells. Following parasite growth and division the host cell lyses, releasing less than 20 B. exitiosa which are in turn phagocytosed. Increased haemocyte production results in a haemocytosis at the expense of gametogenesis. lnfected haemocytes are initially observed in connective tissue, but as infection progresses they can be found in all tissues, with egress via the gonads, kidney, digestive tract, and gill, either by tissue leakage, haemocyte diapedesis, or host decomposition (Hine 1991a). Death may be caused by exhaustion of energy reserves as a result of increasing haemocyte production, rather than a toxic effect produced by the parasite (Hine 1997). Ramilo et al. (2014a) used an in situ hybridisation (ISH) assay to determine that the gonad of O. edulis, was the area where B. exitiosa was most frequently located, and was the exclusive organ of infection in some O. edulis.
Stressors such as exposure to extreme temperatures (7 or 26 °C) and salinity (40%), starvation (prolonged holding in filtered sea water), handling (vigorous stirring four time per day), or heavy infection with an apicomplexan can affect the disease dynamics of B. exitiosa in O. chilensis in New Zealand (Hine et al. 2002, Hine 2002). Co-habitation with infected oysters in holding tanks seems to promote the spread of infection (Hine et al. 2002). Bonamia exitiosa transmission is direct and horizontal, and therefore high densities of oysters in closely spaced beds, and lack of resistance to infection, favour the development of epizootics. However, epizootics decrease stock densities markedly, and are more likely to kill off susceptible oysters rather than those with resistance or tolerance. Thus conditions that do not favour direct horizontal transmission, such as low density and increased resistance, are created, and lower parasite burdens do not cause disease or mortality (Hine 1997).
Mass mortalities due to bonamiasis have not been observed in New Zealand since 1992, and oyster stocks are now recovering, but the parasite is still widespread at low intensity, similar to 1964 levels (Hine and Jones 1994). Lack of mortalities among oysters with low levels of infection may be due to host-parasite kinetics, rather than a change in parasite pathogenicity (Hine 1996a). In Europe, B. exitiosa was associated with O. edulis mortality in Galicia, Spain (Carrasco et al. 2012). However, low prevalence of infection and no mortalities were reported with the detection of B. exitiosa in O. edulis on the Adriàtic coast of Italy (Narcisi et al., 2010) and on the Mediterranean coast of Spain (Carrasco et al. 2012) nor in O. stentina on the coast of Tunisia (Hill et al., 2010). In San Matías Gulf, Argentina, where a mortality rate of 95% was recorded in cultured Ostrea puelchana in 1995-1996, assessment of the status of B. exitiosa in natural populations of O. puelchana 14 years after the epizootic indicated that oyster density was markedly decreased and oyster mean sizes was lower, indicating that bonamiosis controlled the population structure of the oysters in the persisting oyster beds during this period (Oehrens Kissner et al. 2014).
In an attempt to understand the molecular basis of the immune response of O. edulis against bonamiosis, Martín-Gómez et al. (2012) used a combination of suppression subtractive hybridization and Quantitative-Polymerase Chain Reaction (q-PCR) approaches to identify genes involved in the development of responses to infection both in early and advanced phases of the disease caused by B. exitiosa and/or Bonamia ostreae. They determined that the expression of a number of genes related with signal transduction, oxidative stress, chaperones and with leukotriene synthesis or inflammation in O. edulis haemocytes changed in association with bonamiosis (Martín-Gómez et al. 2012).
Diagnostic techniques
Engelsma and Hine (2009a) indicated that PCR, heart imprints, and histology can be used for targeted surveillance and presumptive diagnosis, and DNA sequencing is the recommended method for confirmatory diagnosis. Engelsma et al. (2014) presented a summary and discussion of the various diagnostic techniques to detect Bonamia spp.
Gross Observations
Engelsma and Hine (2009a) indicated that B. exitiosa in O. angasi in Australia caused large focal necrotic lesions. However, in O. chilensis in New Zealand infections were disseminated throughout the oyster and there were no gill lesions although congested gills may burst (Engelsma and Hine 2009a).
Tissue Imprint (“Heart Smears”)
Make acetone- (or methanol-) fixed impression smears from heart tissue (preferably ventricle since the auricles contain an abundance of serous cells which make detection of the parasite difficult). Stain with Wright, Wright-Giemsa or equivalent stain (e.g. Hemacolor, Merck; Diff-QuiK, Baxter). Examine for 2-5 µm spherical or ovoid organisms with a central nucleus (fried egg appearance) within or outside the haemocytes. Note: The organisms are enlarged by this method compared to those in fresh or histological preparations. Diggles et al. (2003) determined that the examination of stained heart imprints appears to be the most time and cost effective method for screening large numbers of oysters in situations where reduced sensitivity may be tolerated but high specificity (reduced diagnosis of false positive results) is required.
Histology
Examine haematoxylin and eosin stained tissue cross-sections for haemocyte inclusions in the 2-3 µm size-range (Moore 2006). Bonamia exitiosa is distributed systemically in advanced infections. In early infections, B. exitiosa is often observed within haemocytes in focal infiltrations in the connective tissue of the gill and mantle, and in the vascular sinuses around the stomach and intestine. Histology appears most useful in epidemiological studies where detection of physiological state, other associated disease agents or pathological lesions is required (Diggles et al. 2003). Light microscopy (histology and tissue imprints) cannot be used to distinguish between species of Bonamia (Diggles et al. 2003, Engelsma and Hine 2009a). However, in Ostrea edulis infected with both B. exitiosa and B. ostreae, Abollo et al. (2008) found B. exitiosa to be larger (mean diameter = 2.8 µm, SE = 0.07 µm, range: 2–5 µm, N = 73) with a central nucleus (sometimes subcentral but rarely peripheral), compared to the smaller B. ostreae (mean diameter = 1.6 µm, SE = 0.04 µm, range: 1–2.5 µm, N = 55) with a peripheral nucleus and scant cytoplasm. Dungan et al. (2012) reported that the largest intra-haemocytic B. exitiosa in C. ariakensis from Florida had diameters of 3 μm with central acidophilic nuclei (0.5 μm in diameter) and plasmodium-like bodies containing numerous, small nuclei or cells also infected circulating haemocytes. Similarily, Longshaw et al. (2013) described the B. exitiosa in O. edulis from England as measuring approximately 2 to 3 μm in diameter with a central or subcentral nucleus with infected haemocytes containeingd up to five microcells, and rarely, larger plasmodia-like multinucleated stages characterised by irregular shape and increased eosinophilic cytoplasm were noted in the haemocyte cytoplasm.
Bonamia exitiosa can cause very different histopathology in different hosts (Engelsma and Hine 2009a). For example, in O. angasi in Australia, it occurs in focal gill lesions, thought to be the point of entry, and low numbers of parasites are observed in large focal necrotic lesions. The parasite is epitheliotropic. In O. chilensis in New Zealand infections are disseminated throughout the oyster, there are no gill lesions, although congested gills may burst. Large numbers of parasites may be present, and the parasite is not epitheliotropic (Engelsma and Hine 2009a). Longshaw et al. (2013) indicated that haemocyte infiltration of the connective tissues surrounding the digestive gland and the mantle along with necrosis of the tissues was associated with the infection of B. exitiosa in O. edulis from England. However, as indicated by Hill et al. (2010b), histopathology is not a suitable tool for species identification because observed differences could be host mediated.
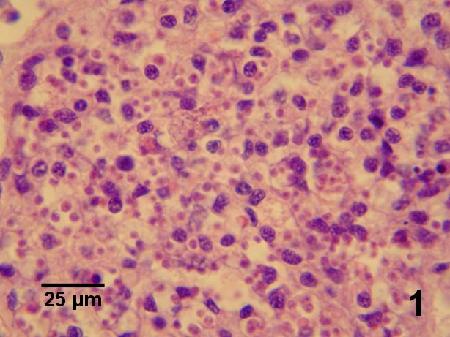
Figure 1. Heavy infection of Bonamia exitiosa within haemocytes in the gonad of Ostrea chilensis (see Fig. 2 for higher magnification). Image provided by Ben Diggles, Ph. D., DigsFish Services.
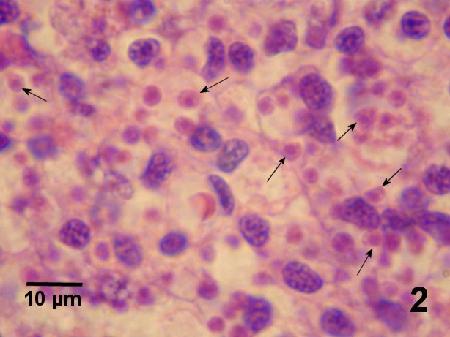
Figure 2. Magnification of Bonamia exitiosa (arrows) from Fig. 1. Image provided by Ben Diggles, Ph. D., DigsFish Services.
Electron Microscopy
Prepare tissues according to standard procedures for electron microscopy (e.g., Moore 2006). The ultrastructure of B. exitiosa resembles other haplosporidians in the possession of haplosporosomes, haplosporogenesis, persistence of mitotic microtubules during interphase and of the nuclear envelope during mitosis, and occurrence of a diplokaryotic or multi-nucleate plasmodial stage. Unlike other haplosporidians, a stage containing a large vacuole derived from enlargement of one or more mitochondria has been observed in B. exitiosa. Plasmodial forms of B. exitiosa are distinguished from other developmental stages of this parasite by their size (4.0-4.5 µm), irregular cellular and nuclear outline, amorphous cytoplasmic inclusions (multi-vesicular bodies), and arrays of Golgi-like smooth endoplasmic reticulum. Forms of intermediate cytoplasmic density were more electron dense than the plasmodial forms and slightly smaller in diameter (3.0-3.5 µm). Haplosporosomes are formed from Golgi/nuclear material complexes and are similar in construction and structure to some viruses. Hine and Wesney (1992, 1994a) hypothesized that the haplosporosomes may represent an early viral element in eukaryotic cells. Hine et al. (2014) regarded metabolic features (such as mitochondrial profiles, lipid droplets and endoplasmic reticulum) as having little taxonomic value. Because ultrastructure features associated with the process of haplosporogenesis (including Golgi, indentations in the nuclear surface, the putative trans-Golgi network, perinuclear granular material and haplosporosome-like bodies) are not understood, only haplosporosome shape and size may be of some taxonomic value (Hine et al. 2014).
Hill et al. (2010b) indicated that ultrastructure can be used to differentiate between B. exitiosa and B. ostreae. However, they cautioned that care must be taken to compare equivalent stages as these two species have simple uni-nucleate stages that become bi-nucleate and divide into further uni-nucleate stages, and some observed differences can be host defined. Nevertheless, B. exitiosa has a large amoeboid trophic stage that is apparently lacking in B. ostreae. Also, the simple uni-nucleate stage of B. ostreae is smaller (2.4 ± 0.5 μm) than B. exitiosa (3.0 ± 0.3 μm), B. ostreae has fewer and larger haplosporosomes (7 ± 5, 153 ± 18 nm) than B. exitiosa (14 ± 6, 148 ± 11 nm), and B. ostreae has fewer mitochondrial sections (2 ± 1) than B. exitiosa (3 ± 1) (Hill et al. 2010b). Dense forms of the uni-nucleate stage are also different in the two species. Dense forms of B. exitiosa are less dense; slightly larger in size (3.0 ± 0.3 µm mean diameter n = 61 in comparison to B. ostreae with a mean diameter of 2.4 ± 0.5 µm, n = 64); have more haplosporosomes, mitochondrial profiles and lipoid bodies per ultrastructure section; and have smaller tubulovesicular mitochondria than B. ostreae. In addition, dense forms of B. ostreae lack nuclear membrane-bound Golgi/nuclear cup complexes and a vacuolated stage (Hine et al. 2001). Despite these ultrastructural differences between B. ostreae and B. exitiosa as described by Dinamani et al. (1987), Hine et al. (2001) and Hill et al. (2010b); Narcisi et al. (2010) found that the ultrastructural characteristics of B. exitiosa occurring in Italy were so variable that they cannot be used to definitively identify a species of Bonamia. Nevertheless, Hine et al. (2014) indicated that the ultrastructure of Bonamia in O. angasi from Australia, Crassostrea ariakensis from the USA, O. puelchana from Argentina and O. edulis from Spain all appear conspecific with B. exitiosa.
DNA Probes
Molecular procedures for the analysis of samples for Bonamia spp. were described by Moore (2006) and various other probes used in similar procedures are described by the various authors indicated below. The sequence of segments of the ribosomal RNA locus which includes parts of the small subunit ribosomal DNA (SSU rDNA or 18S rDNA) gene, the internal transcribed spacer (ITS) regions (consisting of ITS1-5.8S-ITS2) and the large subunit ribosomal DNA (LSU rDNA) gene of B. exitiosa have strong identity with that of B. ostreae but is sufficiently divergent to show polymorphism by restriction fragment length polymorphism (RFLP) analysis (by digesting polymerase chain reaction (PCR) product (amplified with primers BO and BOAS as described by Cochennec et al. (2000)) with Bgl1 (Promega)). The B. ostreae profile consisted of 2 bands of 120 and 180 base pairs (bp) while the B. exitiosa profile consisted of a unique band of 304 bp (Hine et al. 2001). The differences between B. exitiosa and B. ostreae was verified by the analysis of DNA sequences from this region of the genome (Hill et al. 2010b). In addition, sequence analysis of this region of the genome was used to confirm that the Bonamia isolates from O. edulis on the Galician coast (NW Spain), the Ebro Delta on the Mediterranean coast of Spain, the Manfredonia Gulf (Adriatic Sea), Italy and southwest coast of England, from O. chilensis in New Zealand, from C. ariakensis in Florida and North Carolina, USA, and from Saccostrea glomerata and Ostrea angasi in Australia were B. exitiosa (Abollo et al. 2008, Narcisi et al. 2010, Hill et al. 2010b, Carrasco et al. 2012, Dungan et al. 2012, Longshaw et al. 2013, Ramilo et al. 2014b, Carnegie et al. 2014).
Although the in situ hybridisation (ISH) assay outlined by Cochennec et al. (2000) also detected other species of haplosporidians (e.g., Haplosporidian nelsoni, see Carnegie and Cochennec-Laureau 2004), it appeared to be a better technique than PCR for screening small numbers of oysters when high sensitivity is required and when strict control over fixation and screening is possible (Diggles et al. 2003). For reliable ISH results, Diggles et al. (2003) recommended that samples not be fixed for longer than 48 hr in formalin (10% formalin in sea water) nor be transferred to 70% ethanol for prolonged periods of time after fixation. The optimisation of PCR for the specific detection of B. exitiosa will provide a sensitive diagnostic technique but other visual confirmation methods such as ISH must be employed to rule out the possibility of false positives (Diggles et al. 2003). Hill et al. (2010a) used a cocktail of 3 probes and Ramilo et al. (2014a) also designed oligonucleotide probes for the specific detection of B. exitiosa in ISH assays which were species-specific and more sensitivie than traditional histology to visualise the parasite inside host tissue.
A real-time TaqMan PCR assay was developed for the detection of Bonamia spp. that was more sensitive than histology, was comparable to conventional PCR in sensitivity but produced more rapid results with a low risk of sample cross-contamination, and could be optimised to determine the intensity of infection (Corbeil et al. 2006a). Ramilo et al. (2013) described species-specific conventional PCR (cPCR) and real-time PCR diagnostic assays for B. exitiosa and B. ostreae in O. edulis as well as a multiplex PCR method to detect both parasites in a single assay. The sensitivity of these procedures was higher using oyster gills and gonad tissue, rather than gills alone. Although the implementation of statistical tools (maximum likelihood method) for the comparison of these assays and histology showed the possibility of false positives, all procedures showed negative results when used for the analysis of oysters from a Bonamia-free area (Ramilo et al. 2013).
Methods of control
To date there are no known eradication or control procedures. Effective management of the disease caused by B. exitiosa is complicated by the extensive nature of the oyster production process and limited options for disease control of the cultured stocks in open water surrounded by wild oyster populations (Engelsma et al. 2014). Hine (1996a) indicated that B. exitiosa can be transmitted directly from oyster to oyster and therefore disease spread is related to stocking density. Until the method(s) of transmission and host specificity of this parasite has been fully investigated, the movement of oysters out of endemic areas should be avoided. For example, Hill et al. (2010b) speculated that the B. exitiosa-like organism reported by Bishop et al. (2006) from Morehead City Bay, North Carolina, USA was possibly a shipping introduction. Also, the initial mortalities in San Antonio Este, Argentina, occurred in a bed 1 km from an international shipping wharf, suggesting that the associated B. exitiosa outbreak could have originated from a shipping introduction (Hill et al. 2010b).
The apparent reduction in pathogenicity and/or selection for B. exitiosa tolerant stocks may explain the 20 to 30 year cycles of large mortalities experienced by O. chilensis in Foveaux Strait, New Zealand. Cranfield et al. (2005) proposed that mechanical disturbances of oysters by increasingly intense dredging and resulting modification of the benthic habitat was a major source of stress that correlated with the epizootic. They indicated that the recovery of the oyster population was closely linked to the regeneration of habitat. They also suggested that the fishery could be improved by mitigating mechanical disturbances during dredging by use of lighter dredges and less damaging towing strategies as well as pursuing rotational fishing strategies that allow benthic habitat to regenerate from undisturbed areas (Cranfield et al. 2005).
References
Abollo, E., A. Ramilo, S.M. Casas, P. Comesaña, A. Cao, M.J. Carballal and A. Villalba. 2008. First detection of the protozoan parasite Bonamia exitiosa (Haplosporidia) infecting flat oyster Ostrea edulis grown in European waters. Aquaculture 274: 201–207.
Arzul, I., A. Langlade, B. Chollet, M. Robert, S. Ferrand, E. Omner, S. Lerond, Y. Couraleauy, J.P. Joly, C. François and C. Garcia. 2011. Can the protozoan parasite Bonamia ostreae infect larvae of flat oysters Ostrea edulis? Veterinary Parasitology 179: 69-76.
Audemard, C., R.B. Carnegie, N.A. Stokes, M.J. Bishop, C.H. Peterson and E.M. Burreson. 2008a. Effects of salinity on Bonamia sp. survival in the Asian oyster Crassostrea ariakensis. Journal of Shellfish Research 27: 535-540.
Audemard, C., R.B. Carnegie, M.J. Bishop, C.H. Peterson and E.M. Burreson. 2008b. Interacting effects of temperature and salinity on Bonamia sp. parasitism in the Asian oyster Crassostrea ariakensis. Journal of Invertebrate Pathology 98: 344-350.
Audemard, C., R.B. Carnegie, K.M. Hill, C.H. Peterson and E.M. Burreson. 2014. Bonamia exitiosa transmission among, and incidence in, Asian oyster Crassostrea ariakensis under warm euhaline conditions. Diseases of Aquatic Organisms 110: 143-150.
Berthe, F.C.J. and P.M. Hine. 2003. Bonamia exitiosa Hine et al., 2001 is proposed instead of B. exitiosus as the valid name of Bonamia sp. infecting flat oysters Ostrea chilensis in New Zealand. Diseases of Aquatic Organisms 57: 181.
Bishop, M.J., R.B. Carnegie, N.A. Stokes, C.H. Peterson and E.M. Burreson. 2006. Complications of a non-native oyster introduction: Facilitation of a local parasite. Marine Ecology Progress Series 325: 145-152.
Burreson, E.M., N.A. Stokes and R.B. Carnegie. 2004. Bonamia sp. (Haplosporidia) found in nonnative oysters Crassostrea ariakensis in Bogue Sound, North Carolina. Journal of Aquatic Animal Health 16: 1-9.
Campalans, M., P. Rojas and M. Gonzalez. 2000. Haemocytic parasitosis in the farmed oyster Tiostrea chilensis. Bulletin of the European Association of Fish Pathologists 20: 31-33.
Carnegie, R.B. and N. Cochennec-Laureau. 2004. Microcell parasites of oysters: recent insights and future trends. Aquatic Living Resources 17: 519-528.
Carnegie, R.B. and M.Y. Engelsma. 2014. Microcell parasites of molluscs: introduction to DAO Special 7. Diseases of Aquatic Organisms 110: 1-4.
Carnegie, R.B., N.A. Stokes, C. Audemard and E.M. Burreson. 2005. Bonamiasis in the crested oyster Ostrea equestris in North Carolina, USA. Journal of Shellfish Research 24: 644. (Abstract).
Carnegie, R.B., E.M. Burreson, P.M. Hine, N.A. Stokes, C. Audemard, M.J. Bishop and C.H. Peterson. 2006. Bonamia perspora n. sp. (Haplosporidia), a parasite of the oyster Ostreola equestris, is the first Bonamia species known to produce spores. Journal of Eukaryotic Microbiology 53: 232-245.
Carnegie, R.B., N.A. Stokes, C. Audemard, M.J. Bishop, A.E. Wilbur, T.D. Alphin, M.H. Posey, C.H. Peterson and E.M. Burreson. 2008. Strong seasonality of Bonamia sp. infection and induced Crassostrea ariakensis mortality in Bogue and Masonboro Sounds, North Carolina, USA. Journal of Invertebrate Pathology 98: 335-343.
Carnegie, R.B., K.M. Hill, N.A. Stokes and E.M. Burreson. 2014. The haplosporidian Bonamia exitiosa is present in Australia, but the identity of the parasite described as Bonamia (formerly Mikrocytos) roughleyi is uncertain. Journal of Invertebrate Pathology 115: 33-40.
Carrasco, N., A. Villalba, K.B. Andree, M.Y. Engelsma, B. Lacuesta, A. Ramilo, I. Gairín and M.D. Furones. 2012. Bonamia exitiosa (Haplosporidia) observed infecting the European flat oyster Ostrea edulis cultured on the Spanish Mediterranean coast. Journal of Invertebrate Pathology 110: 307-313.
Cochennec, N., F. LeRoux, F. Berthe and A. Gerard. 2000. Detection of Bonamia ostreae based on small subunit ribosomal probe. Journal of Invertebrate Pathology 76: 26-32.
Corbeil, S., I. Arzul, B. Diggles, M. Heasman, B. Chollet, F.C.J. Berthe and M.S.J. Crane. 2006a. Development of a TaqMan PCR assay for the detection of Bonamia species. Diseases of Aquatic Organisms 71: 75-80.
Corbeil, S., I. Arzul, M. Robert, F.C.J. Berthe, N. Besnard-Cochennec and M.S.J. Crane. 2006b. Molecular characterisation of an Australian isolate of Bonamia exitiosa. Diseases of Aquatic Organisms 71: 81-85.
Cranfield, H.J., A. Dunn, I.J. Doonan and K.P. Michael. 2005. Bonamia exitiosa epizootic in Ostrea chilensis from Foveaux Strait, southern New Zealand between 1986 and 1992. ICES Journal of Marine Science 62: 3-13.
Diggles, B.K., N. Cochennec-Laureau and P.M. Hine. 2003. Comparison of diagnostic techniques for Bonamia exitiosus from flat oysters Ostrea chilensis in New Zealand. Aquaculture 220: 145-156.
Dinamani, P., P.M. Hine and J.B. Jones. 1987. Occurrence and characteristics of the haemocyte parasite Bonamia sp. in the New Zealand dredge oyster Tiostrea lutaria. Diseases of Aquatic Organisms 3: 37-44.
Doonan, I.J., H.J. Cranfield and K.P. Michael. 1994. Catastrophic reduction of the oyster, Tiostrea chilensis (Bivalvia: Ostreidae), in Foveaux Strait, New Zealand, due to infestation by the protistan Bonamia sp. New Zealand Journal of Marine and Freshwater Research 28: 335-344.
Dungan, C.F., R.B. Carnegie, K.M. Hill, C.B. McCollough, S.E. Laramore, C.J. Kelly, N.A. Stokes and J. Scarpa. 2012. Diseases of oysters Crassostrea ariakensis and C. virginica reared in ambient waters from the Choptank River, Maryland and the Indian River Lagoon, Florida. Diseases of Aquatic Organisms 101: 173-183.
Engelsma, M. and M. Hine. 2009a. Infection with Bonamia exitiosa: disease detection, pathogen identification and typing. In: Hill, B., A. Reese, P. Dixon, B. Oidtmann, R. Paley, E. Peeler, G. Stentiford, D. Stone, K. Way, M. Hine, P. Calistri, C. Ippoliti, A. Di Lorenzo, L. Savini, O. Haenen, M. Engelsma (eds), Epidemiology of different agents causing disease in aquatic animals: scientific review and database development, (European Food Safety Authority (EFSA), Parma, Italy), Annex B, pp. 40-41.
Engelsma, M. and M. Hine. 2009b. Infection with Bonamia exitiosa: occurrence and distribution. In: Hill, B., A. Reese, P. Dixon, B. Oidtmann, R. Paley, E. Peeler, G. Stentiford, D. Stone, K. Way, M. Hine, P. Calistri, C. Ippoliti, A. Di Lorenzo, L. Savini, O. Haenen, M. Engelsma (eds), Epidemiology of different agents causing disease in aquatic animals: scientific review and database development, (European Food Safety Authority (EFSA), Parma, Italy), Annex B, 42-44.
Engelsma, M.Y., S.C. Culloty, S.A. Lynch, I. Arzul and R.B. Carnegie. 2014. Bonamia parasites: a rapidly changing perspective on a genus of important mollusc pathogens. Diseases of Aquatic Organisms 110: 5-23.
Hartikainen, H., O.S. Ashford, C. Berney, B. Okamura, S.W. Feist, C. Baker-Austin, G.D. Stentiford and D. Bass. 2014. Lineage-specific molecular probing reveals novel diversity and ecological partitioning of haplosporidians. The International Society for Microbial Ecology Journal (ISME J) 8: 177-186.
Hill, K.M., D.M. White, N.A. Stokes, R.B. Carnegie, N. Aloui-bejaoui, S.C. Webb, P.M. Hine, M.A. Kroeck, R. Ghars alli, R.K. Crockett, T.D. Lewis, K.S. Reece and E.M. Burreson. 2008. New perspectives on the dispersal and evolution of Bonamia species, haplosproidian parasites of oysters. Journal of Shellfish Research 27: 1016. (Abstract).
Hill, K.M., R.B. Carnegie, N. Aloui-Bejaoui, R.E. Gharsalli, D.M. White, N.A. Stokes and E.M. Burreson. 2010a. Observation of a Bonamia sp. infecting the oyster Ostrea stentina in Tunisia, and a consideration of its phylogenetic affinities. Journal of Invertebrate Pathology 103: 179–185.
Hill, B., A. Reese, P. Dixon, B. Oidtmann, R. Paley, E. Peeler, G. Stentiford, D. Stone, K. Way, M. Hine, P. Calistri, C. Ippoliti, A. Di Lorenzo, L. Savini, O. Haenen and M. Engelsma. 2010b. Epidemiology of different agents causing disease in aquatic animals: scientific review and database development (Parma, Italy, European Food Safety Authority (EFSA)), 21 p. Annex E, pp. 109-116.
Hill, K.M., N.A. Stokes, S.C. Webb, P.M. Hine, M.A. Kroeck, J.D. Moore, M.S. Morley, K.S. Reece, E.M. Burreson and R.B. Carnegie. 2014. Phylogenetics of Bonamia parasites based on small subunit and internal transcribed spacer region ribosomal DNA sequence data. Diseases of Aquatic Organisms 110: 33-54.
Hine, P.M. 1991a. The annual pattern of infection by Bonamia sp. in New Zealand flat oysters, Tiostrea chilensis. Aquaculture 93: 241-251.
Hine, P.M. 1991b. Ultrastructural observations on the annual infection pattern of Bonamia sp. in flat oysters Tiostrea chilensis. Diseases of Aquatic Organisms 11: 163-171.
Hine, P.M. 1992. Ultrastructural and ultracytochemical observations on Bonamia sp. in oysters (Tiostrea chilensis), with a consideration of organelle function. Aquaculture 107: 175-183.
Hine, P.M. 1996a. The ecology of Bonamia and decline of bivalve molluscs. New Zealand Journal of Ecology 20: 109-116.
Hine, P.M. 1996b. Southern hemisphere mollusc diseases and an overview of associated risk assessment problems. Revue Scientifique et Technique de l'Office International des Epizooties 15: 563-577.
Hine, P.M. 1997. Health status of commercially important molluscs in New Zealand. Surveillance 24: 25-28.
- Date modified: